PhosphoSCOPE
SCoPE-MS is a proteomics method that allows for the analysis of single mammalian cells by mass spectrometry. Read the open-access article here, in Genome Biology:
https://genomebiology.biomedcentral.com/articles/10.1186/s13059-018-1547-5
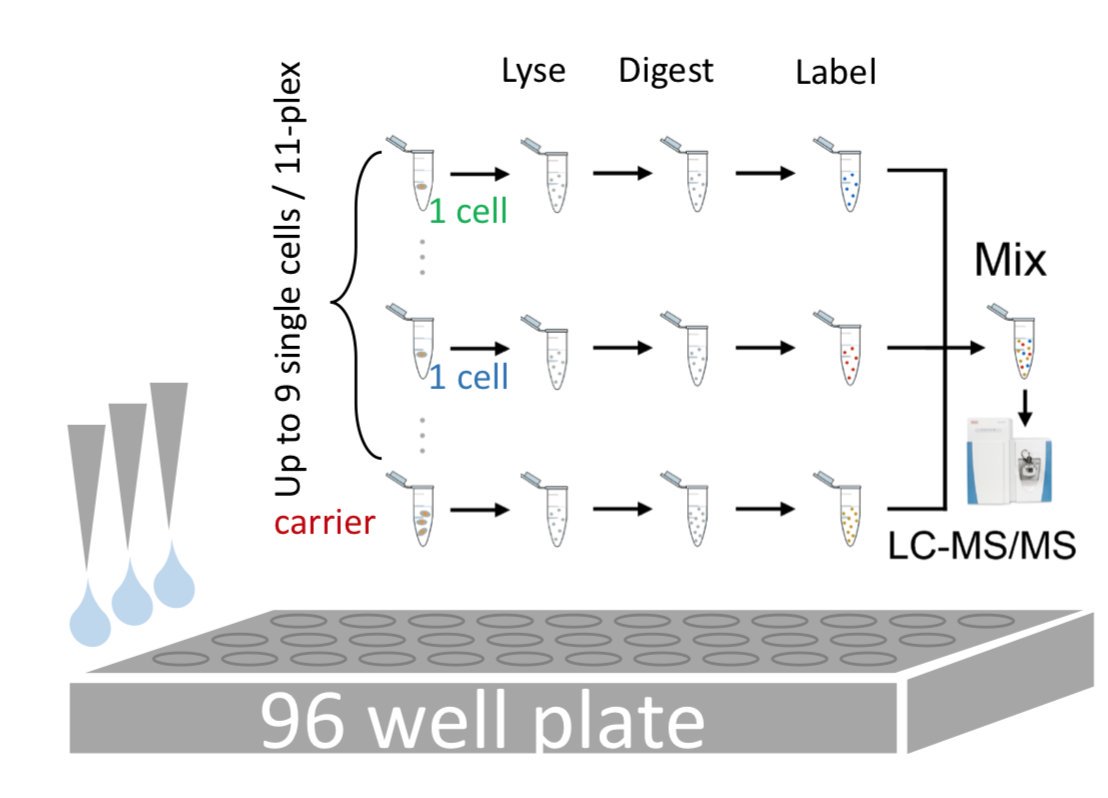
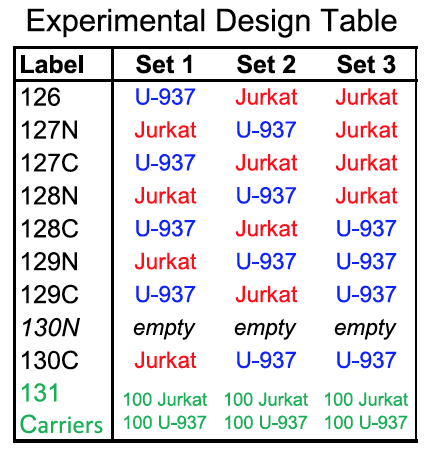
I joined the Slavov Lab in the middle of the development of SCoPE-MS, and was able to contribute by developing the last part of the quantitation pipeline – normalizing and filtering our data from MaxQuant so that it could be used for downstream biological analyses. I also helped clean up our core MATLAB processing code.
My more recent work with SCoPE-MS has been leading a project that applies its concepts to other areas of proteomics. For me this involves:
- Mammalian cell culture, mainly on blood cancer cell lines Jurkat (T-cells) and U-937 (monocytes)
- Proteomics sample preparation:
- mPOP cell lysis protocols – available here on bioRXiV
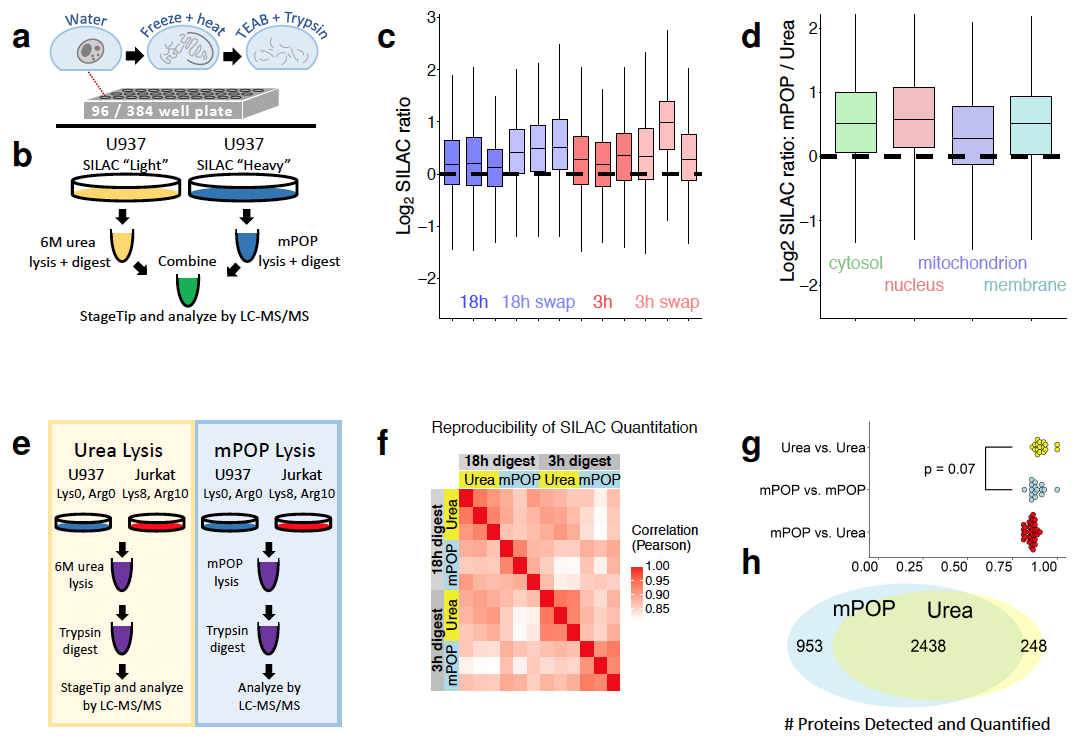
Figure 1. from Specht et al 2018
-
- Experimental cell lysis protocols
- Conventional and experimental protein digestions
- Various cleanup procedures, such as with C18 StageTips or SPE with Waters SepPaks
- Labeling, and optimizing labeling, with Thermo tandem-mass-tags (TMT)
- Running a variety of MS quantitation methods
- Isobaric-tag, TMT
- SILAC
- Label-free/iBAQ
- Optimizing LC-MS parameters
- Current system is with a Dionex LC, and a base model Q-Exactive.
- Front end/chromatography is changing frequently, but it’s usually a standard reverse phase, nano-LC column, with a fused ESI spray tip and an ABIRD in the source.

Our lab’s setup with a shrine to Alexander Makarov, inventor of the Orbitrap analyzer
In addition to running physical experiments I wrote software tools to help process our data. These are written in a mix of R, python, and MATLAB.
Much of our analysis has been offloaded/automated with the DO-MS application, which I encourage you to check out! It eliminates the process of creating scripts for each individual experiment, and allowed me to quickly share reports with my colleagues.
For more information, see my poster: P-SCOPE: Carrier-enabled low-input phosphoproteomics by mass spectrometry
← Back to home